DNA methylation is a broadly observed epigenetic modification in living systems, playing diverse functions in transcriptional regulation, transposable element silencing, as well as innate immunity.
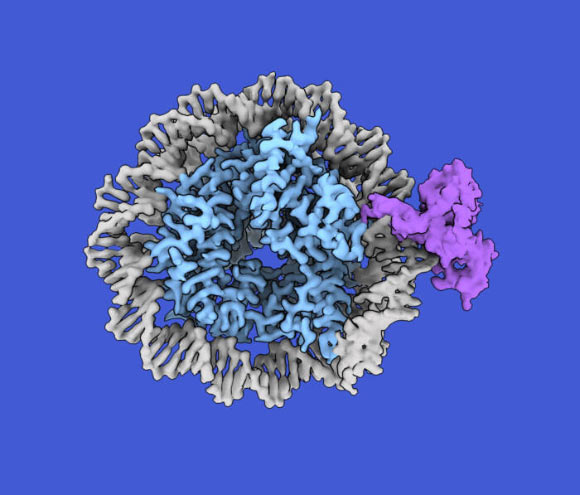
A nucleosome composed of DNA (gray) and histones (blue) with a single hemimethylated cytosine bound by CDCA7 (purple). Image credit: Kyohei Arita / Yiming Niu.
DNA methylation is a process in which a methyl group is attached to the cytosine base of the DNA molecule, and a major way that DNA is epigenetically marked.
Epigenetic modifications can act as on-off switches to regulate gene expression and help generate diverse cell types without changing the underlying DNA sequence. It is how the body ensures that brain-related genes don’t get turned on in heart cells, for example.
For this reason, maintenance of the DNA methylation pattern is important to ensure the correct and consistent function of each cell type.
But this is no easy feat: the DNA methylation pattern can change over time, and this is linked to a variety of diseases.
One is a rare genetic condition called immunodeficiency, centromeric instability and facial anomalies (ICF) syndrome, whose symptoms include recurrent respiratory infections, facial anomalies, and slowed growth and cognition.
While it has been known that mutations in the CDCA7 gene cause ICF syndrome, little was known about the gene’s molecular function.
In new research, Rockefeller University Professor Hironori Funabiki and his colleagues identified a unique functional feature of CDCA7 that ensures the accurate inheritance of DNA methylation.
The researchers discovered that CDCA7 senses hemimethylation in eukaryotes — an important find, because hemimethylation sensing has long been thought to be solely carried out by a protein called UHRF1.
“It’s quite an incredible finding,” said Rockefeller University scientist Isabel Wassing.
“Learning that CDCA7 also acts as a sensor explains why its mutation leads to diseases like ICF syndrome and fills in a major gap in the field of epigenetics.”
“But it also introduced new questions. Why, for example, does the cell need two different hemimethylation sensors?”
“We found that the CDCA7 gene, known as the causative gene of ICF syndrome, promotes DNA methylation by controlling the ubiquitination of histone H3 through specific binding to hemimethylated DNA on nucleosomes,” said University of Tokyo researcher Atsuya Nishiyama.
Scientists know that the access of many enzymes and DNA-binding proteins is restricted by chromatin, including those that are necessary to introduce methylation to the DNA.
Earlier research by Professor Funabiki’s team showed that CDCA7 forms a complex with the protein encoded by the HELLS gene, whose mutations also cause ICF syndrome.
HELLS is a so-called nucleosome remodeler, which can temporarily unwrap the DNA molecule from the nucleosome.
“We envisioned that the CDCA7-HELLS complex is important to help the cell overcome the barrier of compacted heterochromatin and make the DNA molecule accessible to the deposition of methylation,” Professor Funabiki said.
“But there are many different nucleosome remodelers that are able to expose the DNA molecule in this way.”
“It remained a mystery to us why CDCA7-HELLS is the only nucleosome remodeling complex directly associated with DNA methylation maintenance.”
“Now that we’ve shown that CDCA7 specifically recruits HELLS to hemimethylated DNA, this finally provides an explanation.”
In this model, CDCA7 recognizes the hemimethylated DNA in chromatin and recruits HELLS to the site, which, as a nucleosome remodeler, slides the nucleosome out of the way, revealing the hemimethylation site to UHRF1.
The handover of hemimethylation sensing indicates that CDCA7 is better at detecting hemimethylation within the dense heterochromatin than UHRF1 is. It also explains the cell’s need for two different sensors.
“For these sensors to detect hemimethylation, they must directly and selectively bind the hemimethylated DNA substrate,” Dr. Wassing said.
“CDCA7 seems uniquely able to do that while the DNA is wrapped around the nucleosome. Without it, UHRF1 would be blind to the hemimethylation signal within the nucleosome particles.”
“Our findings suggest that CDCA7 and HELLS promote DNA methylation in a mechanism distinct from de novo DNA methylation, which is now consolidated by our demonstration that the CDCA7 HMZF domain specifically recognizes hemimethylated CpG, the substrate of the maintenance DNA methyltransferase DNMT1,” Dr. Nishiyama said.
“ICF disease-associated mutations in the CDCA7 gene abolish its hemimethylated DNA binding, supporting the functional importance of hemimethylation detection by CDCA7.”
This new understanding may help illuminate the underlying mechanisms of diseases born from dysfunctional methylation.
In the future they’ll seek out functions for hemimethylation sensors beyond DNA methylation maintenance.
“Since some chromosomal regions are known to preserve hemimethylation status, their recognition by CDCA7 may have much broader roles in gene regulation and chromosome organization. It’s an exciting possibility,” Professor Funabiki said.
“Our study lays the groundwork for the development of new DNA methylation inhibitors and therapeutic drugs for ICF syndrome,” Dr. Nishiyama said.
“Therapies that artificially regulate CDCA7-dependent DNA methylation may also prevent cancer and aging and help extend healthy lifespan.”
The findings appear this month in the journal Science Advances.
_____
Isabel E. Wassing et al. 2024. CDCA7 is an evolutionarily conserved hemimethylated DNA sensor in eukaryotes. Science Advances 10 (34); doi: 10.1126/sciadv.adp5753
This article is based on a press-release from Rockefeller University.